MCRiceRepGP
Parameters
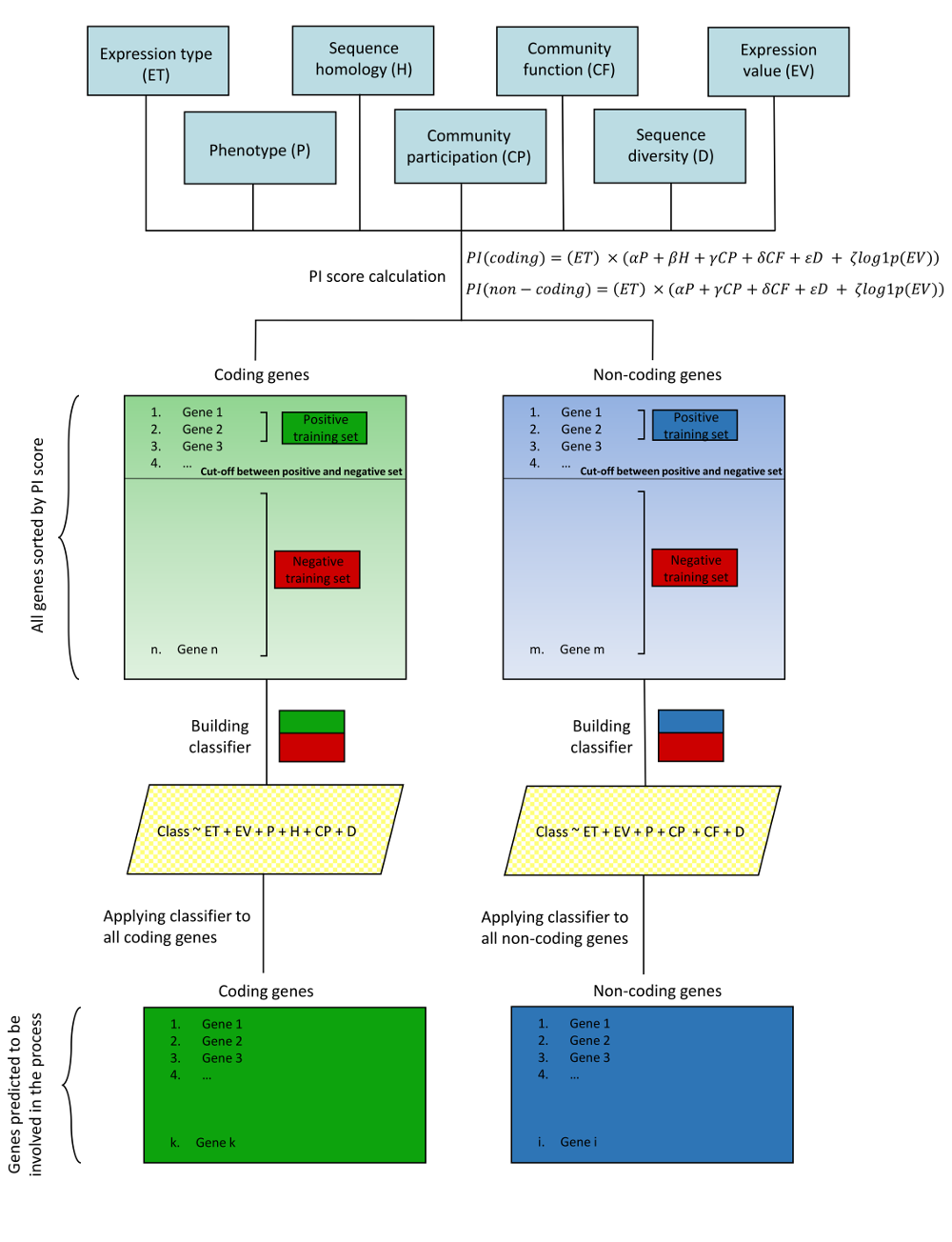
Expression type (ET, value = 0 or 1) - Is the highest expression recorded in chosen tissue type (rep - reproductive or veg - vegetative).
Phenotype category (P, value = 0 or 1) - Is the phenotype category consistent with tissue type.
Sequence homology (H, value = 0 or 1) - Is the homolog annotated with functions found among key words.
Community participation (CP, value = 0 or 1) - Is the gene found within a community in co-expression network.
Community function (CF, value = 0 or 1) - Does the gene belong to a community annotated with functions found among the key words.
Sequence diversity (D, value = 0 or 1) - Does the gene display low sequence diversity within the species.
Expression value (EV, value = log(FPKM)) - The FPKM value for the gene in the tissue with highest expression.
The weights of contribution of P, H, CP, CF, D and EV to the PI score can be adjusted.
The features included in the classifier (ET, P, H, CP, CF, D and EV) can be adjusted.
Notes
Please be mindful that the final gene set identified by MCRiceRepGP will reflect the positive and negative training sets.
The genes are ranked by PI score. Always inspect the final output, especially bottom of the list. For example, in some cases
the genes will be included only on the basis on CP and EV. This is expected behaviour, which can be changed by lowering CP
weight in PI score and removing the CP feature from classifier. Also please ensure a level of compatibility between PI score
weights and features selected for classifier.
Download
Files included in download: Results table, fasta sequences, training sets, classifier, summary of settings.
Community annotation
Annotation of communities is too large to be displayed in the browser.
Please download MCRiceRepGP.rice.files.11012018.tgz from https://osf.io/78axs/ to access community annotation.
Further information
MCRiceRepGP: a framework for identification of sexual reproduction associated coding and lincRNA genes in rice.